Prof. Alexis Vallée-Bélisle’s Scientific Publications
2023
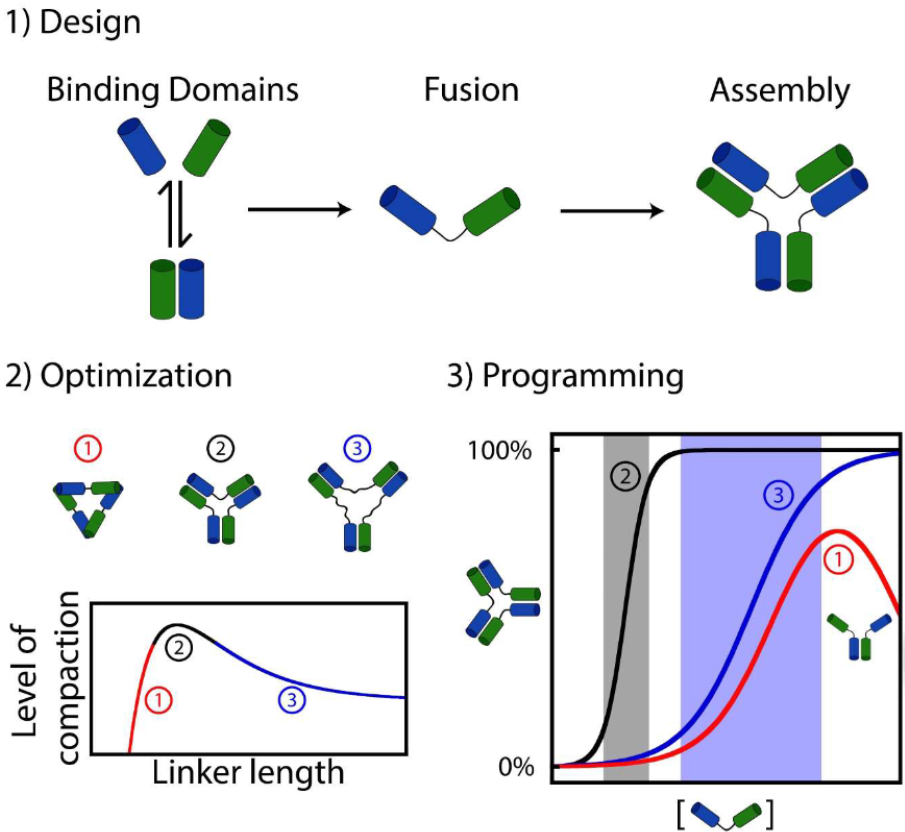
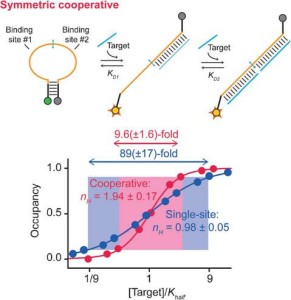
62. Lauzon D & Vallée-Bélisle A*. “Design and Thermodynamics Principles to Program the Cooperativity of Molecular Assemblies” Angew. Chem. e202313944 pdf *corresponding author
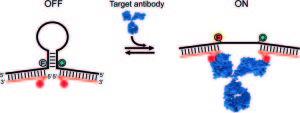
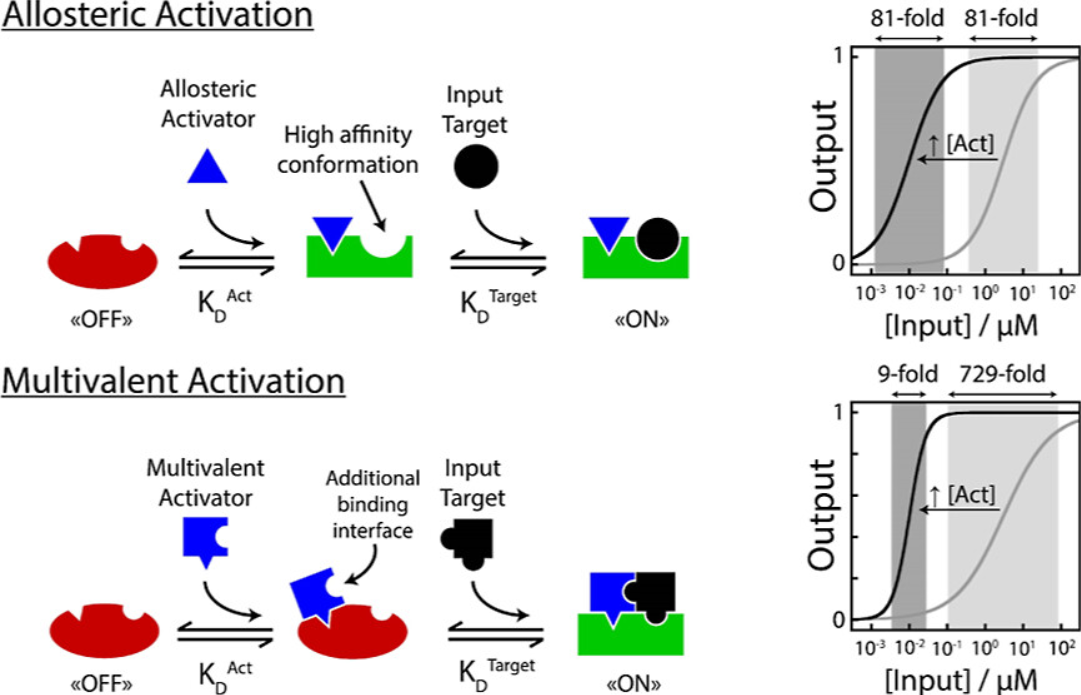
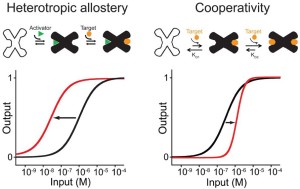
61. Lauzon D & Vallée-Bélisle A*. “Programing Chemical Communication: Allostery vs Multivalent Mechanism” J. Am. Chem. Soc. 145(34):18846–18854. pdf *corresponding author
MSN – 2023-08-15
Molecular communication: Experts are interpreting the language of life
“Canadian researchers at the Université de Montréal have managed to recreate and mathematically validate two crucial molecular languages believed to be at the very core of life itself. […] The research may ultimately expand the horizons of nanotechnology applications, from enhanced biosensing and drug delivery systems to innovative molecular imaging techniques.”
GEN – 2023-08-15
Multivalent Molecular Switches Can Optimize and Program Nanosystems
“Researchers have reconstructed molecular communication interfaces that could aid in advancing nanotechnology development. […] The formation of a multivalent assembly can rationally control the properties of molecular switches, improving their binding efficiency, modifying the drug release properties of DNA cargo, lengthening the residence time of activated switches, or programming DNA-based computation, among many other uses in DNA-based nanotechnologies.”
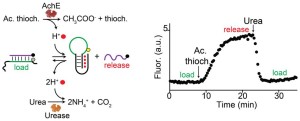
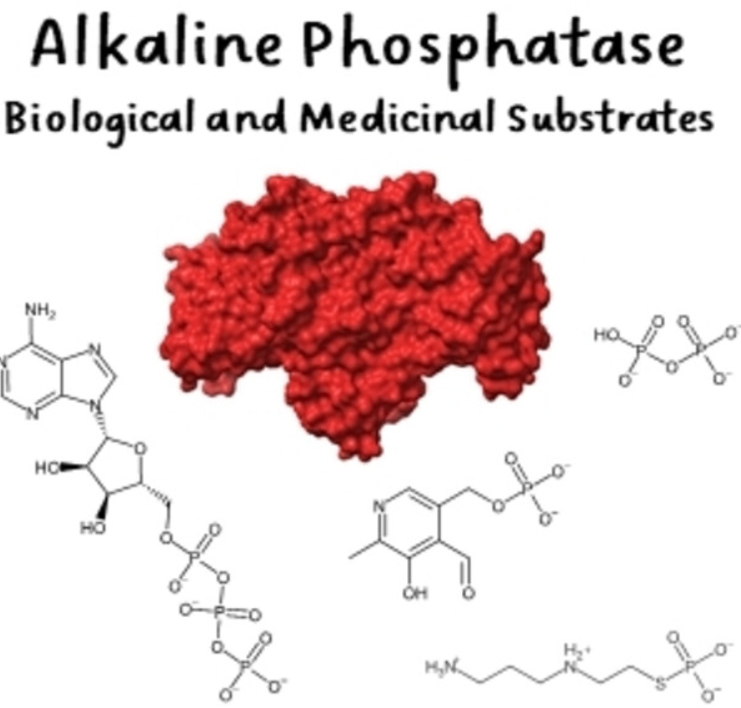
60. Harroun SG* & Vallée-Bélisle A*. “Methods to Characterise Enzyme Kinetics with Biological and Medicinal Substrates: The Case of Alkaline Phosphatase” Chem. Methods e202200067 pdf *corresponding author
59. Lauzon D & Vallée-Bélisle A*. “Functional advantages of building nanosystems using multiple molecular components” Nat. Chem. 15:458–467. pdf *corresponding author
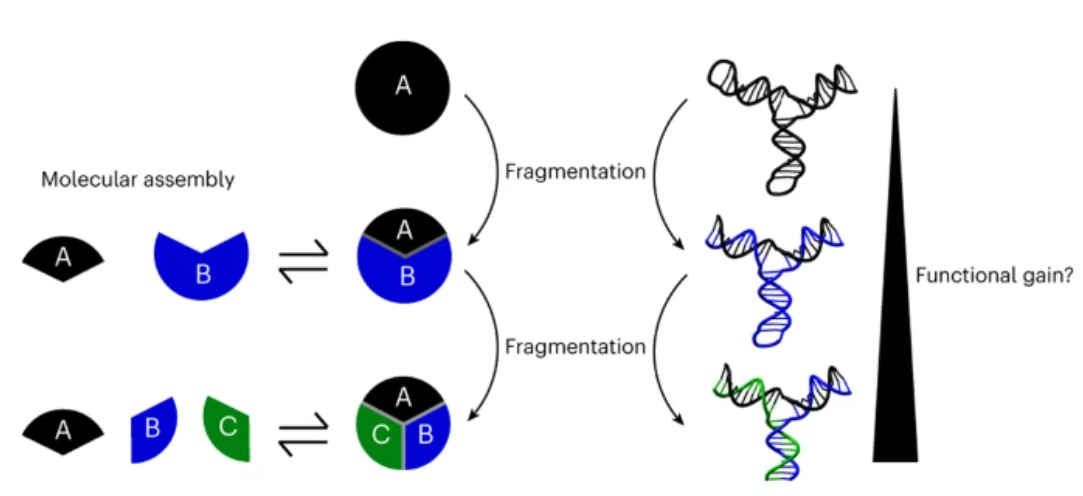
Québec Science – 2023-04-12
Améliorer les nanomachines, un bris à la fois
“Imaginez si briser un objet contribuait à le rendre meilleur. Des chercheurs de l’Université de Montréal détaillent ce mécanisme contre-intuitif découvert dans de minuscules structures biologiques. Lorsqu’un électroménager brise, le mieux qu’on peut espérer est que ses performances ne soient pas trop affectées après la réparation. À l’échelle nanométrique, c’est autre chose : briser et réparer des machines serait non seulement la clé pour une meilleure performance, mais pourrait même leur permettre de développer de nouvelles aptitudes!”
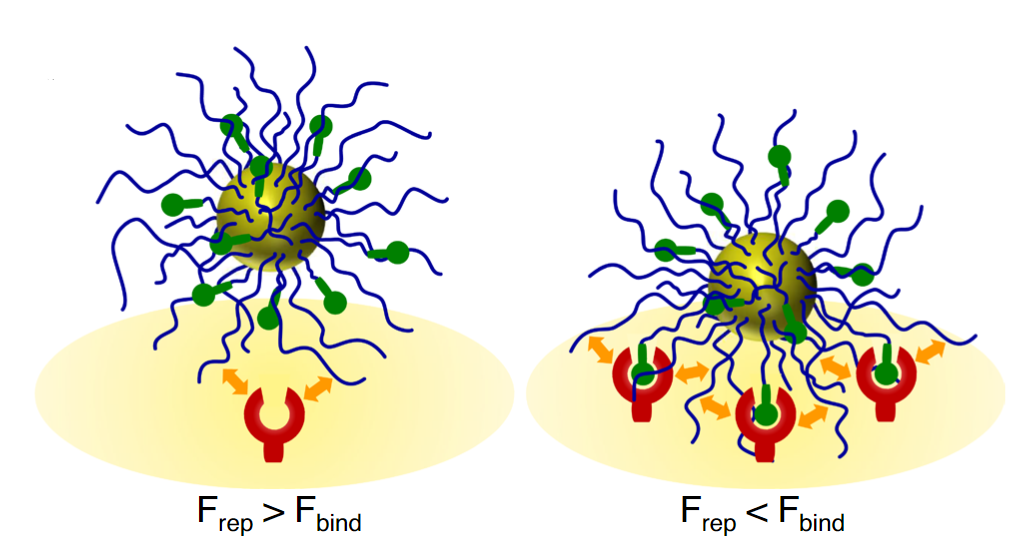
58. Phan HT, Lauzon D, Vallée-Bélisle A, Angioletti-Uberti S, Leblond J, Giasson S. “Bimodal brush-functionalized nanoparticles selective to receptor surface density” Proc. Natl. Acad. Sci. U.S.A. 120(3):e2208377120. pdf
2022
57. Desrosiers A, Derbali RB, Hassine S, Berdugo J, Long V, Lauzon D, De Guire V, Fiset C, DesGroseillers L, Leblond J, Vallée-Bélisle A*. “Programmable self-regulated molecular buffers for precise sustained drug delivery” Nat. Comm. 13:6504. pdf *corresponding author
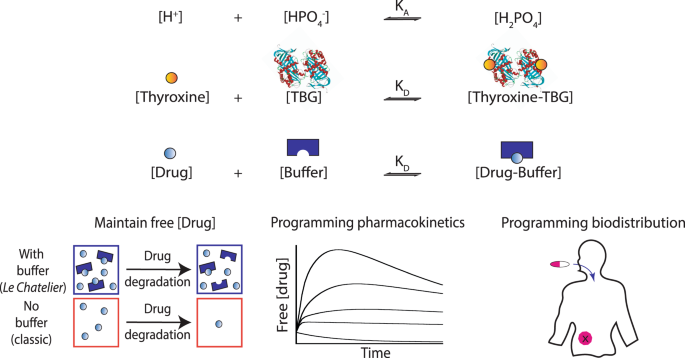
Radio-Canada – 2022-11-10
Des nanotransporteurs d’ADN pour livrer des doses précises de médicaments
“Le dosage n’est pas optimal dans plusieurs traitements contre le cancer. Certains patients reçoivent des doses insuffisantes qui réduisent l’efficacité et peuvent entraîner une résistance aux médicaments, et d’autres reçoivent des doses trop importantes qui causent des effets secondaires. Cette réalité a mené Alexis Vallée-Bélisle et ses collègues à mettre au point des nanostructures s’inspirant de systèmes biologiques qui, eux, réussissent à contrôler et à maintenir la concentration idéale de biomolécules dans le corps.”
News Medical (UK and AUS) – 2022-11-02
Molecular drug transporters could improve treatment of cancers and other diseases
A team of Canadian researchers from Université de Montréal has designed and validated a new class of drug transporters made of DNA that are 20,000 times smaller than a human hair and that could improve how cancers and other diseases are treated. […] One of the key ways to successfully treat disease is to provide and maintain a therapeutic drug dosage throughout treatment. Sub-optimal therapeutic exposure reduces efficiency and typically leads to drug resistance, while overexposure increases side effects. […] UdeM Chemistry associate professor Alexis Vallée-Bélisle, an expert in bio-inspired nanotechnologies, started to explore how biological systems control and maintain the concentration of biomolecules”
2021
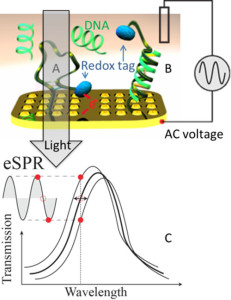
56. Harroun SG, Lauzon D, Ebert M, Desrosiers A, Wang X, Vallée-Bélisle A*. “Monitoring protein conformational changes using fluorescent nanoantennas” Nat. Methods 19(1):71-80. pdf *corresponding author
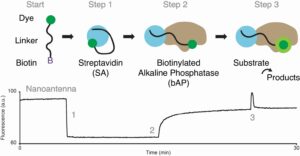
Scientific American – 2022-04-01
Tiny Antennas Made from DNA Light Up Protein Activity
“Cells use protein molecules to communicate with one another and trigger functions throughout the body. When such a message comes into contact with a cell’s surface protein, one of the molecules involved changes shape like a lock opened by a key, prompting a reaction. At just five nanometers across—one 200th the length of a typical bacterium—fluorescent nanoantennas can bind to and interact with proteins on a molecular level. […] For a study in Nature Methods, researchers put these new nanoantennas to work flagging when a particular digestive protein executed five different activities in a solution, such as reacting to antibodies and changing intestinal acidity. “It’s a nice tool in our toolbox,” says the study’s senior author Alexis Vallée-Bélisle, a nanotechnology researcher at the Université de Montréal.”
Science Alert (AUS) – 2022-01-12
Scientists Built The World’s Tiniest Antenna, And It’s Made Out of DNA
“Scientists have built the tiniest antenna ever made – just five nanometers in length. Unlike its much larger counterparts we’re all familiar with, this minuscule thing isn’t made to transmit radio waves, but to glean the secrets of ever-changing proteins. The nanoantenna is made from DNA, the molecules carrying genetic instructions that are around 20,000 times smaller than a human hair. It’s also fluorescent, which means it uses light signals to record and report back information. And those light signals can be used to study the movement and change of proteins in real time. Part of the innovation with this particular antenna is the way in which the receiver part of it is also used to sense the molecular surface of the protein it’s studying. That results in a distinct signal when the protein is fulfilling its biological function.”
Science Magazine (UK) – 2022-01-10
Chemists use DNA to build the world’s tiniest antenna
“Researchers at Université de Montréal have created a nanoantenna to monitor the motions of proteins. Reported this week in Nature Methods, the device is a new method to monitor the structural change of proteins over time – and may go a long way to helping scientists better understand natural and human-designed nanotechnologies. […] Like a two-way radio that can both receive and transmit radio waves, the fluorescent nanoantenna receives light in one colour, or wavelength, and depending on the protein movement it senses, then transmits light back in another colour, which we can detect.”
Radio-Canada – 2022-01-10
Une nanoantenne pour surveiller les mouvements des protéines
“Une antenne à base d’ADN qui permet de surveiller les mouvements des protéines a été mise au point par des scientifiques québécois associés à l’Université de Montréal. Le dispositif de 5 nanomètres de long représente une nouvelle façon de suivre les changements structurels des protéines au fil du temps. […] La mise au point de telles antennes permet d’espérer de nombreuses percées en biochimie et en nanotechnologie. L’enzyme phosphatase alcaline est associée à de nombreuses maladies, dont plusieurs cancers et certaines inflammations intestinales. La création québécoise pourra donc servir à mieux cerner le fonctionnement des protéines, et éventuellement à permettre la création de nouveaux médicaments.”
55. Harroun SG*, Zhang Y, Chen T-Z, Chang H-T*, Vallée-Bélisle A*. “Silver oxide model surface improves computational simulation of surface-enhanced Raman spectroscopy on silver nanoparticles” PCCP 23(29):15480-15484. pdf *co-corresponding author
54. Lauzon D, Guichi Z, Vallée-Bélisle A*. “Engineering DNA Switches for DNA Computing Applications“. In DNA‐ and RNA‐Based Computing Systems; Katz, E., Ed.; WILEY‐VCH GmbH: Weinheim, Germany, 2021; Vol. 1, Chap. 7. pdf

2020
53. Bissonnette S, Del Grosso E, Simon AJ, Plaxco KW, Ricci F*, Vallée-Bélisle A*. “Optimizing the Specificity Window of Biomolecular Receptors Using Structure-Switching and Allostery” ACS Sensors 5(7): 1937-1942. pdf *corresponding author
52. Desrossiers A., Vallée-Bélisle A. “Bio-Inspired DNA Nanoswitches and Nanomachines: Applications in Biosensing and Drug Delivery“. In 21st Century Nanoscience – A Handbook; Sattler, K. S., Ed.; CRC Press: Boca Raton, FL, 2020; Vol. 8, Chap. 6. pdf

2019
51. Mahshid SS, Mahshid S, Vallée-Bélisle A, Kelley SO. “Peptide-Mediated Electrochemical Steric Hindrance Assay for One-Step Detection of HIV Antibodies” Anal. Chem. 91(8):4943-4947. pdf
2018
50. Harroun S, Prévost-Tremblay C, Lauzon D, Desrosiers A, Wang X, Pedro L, Vallée-Bélisle A*. “Programmable DNA switches and their applications” Nanoscale 10(10):4607-4641. pdf * corresponding author
<
49. Wei B, Zhang J, Ou X, Lou X, Xia F, Vallée-Bélisle A*. “Engineering Biosensors with Dual Programmable Dynamic Ranges” Anal. Chem. 90(3):1506-1510. pdf *corresponding author
2017
48. Mahshid SS, Vallée-Bélisle A, Kelley SO. “Biomolecular Steric Hindrance Effects Are Enhanced on Nanostructured Microelectrodes” Anal. Chem. 89(18):9751-9757. pdf
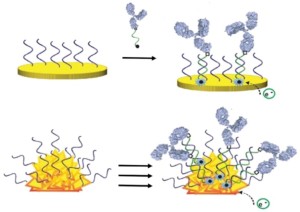
47. Idili A, Ricci F, Vallée-Bélisle A*. “Determining the folding and binding free energy of DNA-based nanodevices and nanoswitches using urea titration curves” Nucleic Acids Res. 45(13):7571-7580. pdf * corresponding author
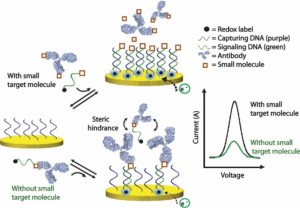
46. Mahshid SS, Ricci F, Kelley SO, Vallée-Bélisle A*. “Electrochemical DNA-Based Immunoassay That Employs Steric Hindrance To Detect Small Molecules Directly in Whole Blood” ACS Sensors 2(6):718−723.pdf *corresponding author
ACS Sens. 2018, 3, 519−519
A Glimpse into the Future of Sensing
“The lectures from the inaugural Advances in Measurement Science Lectureship Awards sponsored by the ACS Analytical Division and ACS Publications were presented at Pittcon on February 27th in Orlando, Florida. (…) Collaborator Alexis Vallée-Bélisle followed on in this theme discussing how the DNA switches could be used to provide insights into different biological protein binding events before discussing some of the electrochemical applications in sensing including his recent paper in ACS Sensors.”
45. Ranallo S, Prévost-Tremblay C, Vallée-Bélisle A, Ricci F. “Antibody-powered nucleic acid release using a DNA-based nanomachine” Nat. Commun. 8:15150. pdf
CBC-News– 2017-05-08
Nanotech ‘slingshot’ shoots drugs right where they’re needed
“A group of Canadian and Italian scientists has developed a nano-scale “slingshot” that can shoot drugs directly to the part of the body that needs them, thereby speeding up recovery and reducing side-effects. This idea addresses one of the most taxing problems in medicine: how to kill diseased cells while preserving healthy ones. Scientists have for many years been working on improving therapies like chemo and radiation on that score, but most efforts have focused on modifying the chemistry rather than altering the delivery of the drug. “It’s all about tuning the concentration of the drug optimally in the body: high concentration where you want it to be active, and low concentration where you don’t want to affect other healthy parts,” says Prof. Alexis Vallée-Bélisle of the University of Montreal, co-author of the report published this week in Nature Communications.”
Science & Vie–2017-05-12
Ce lance-pierre moléculaire envoie les médicaments pile là où il faut
“Ce lance-pierre moléculaire envoie les médicaments pile là où il faut. Prenez un brin d’ADN replié, contenant un médicament. Faites-lui rencontrer un anticorps en forme de Y, synonyme de maladie : un lance-pierre nanométrique va alors se former, propulsant le médicament pile sur sa cible.
Des chimistes italiens et canadiens viennent de créer un lance-pierre à médicament ! L’objectif ? Acheminer et expédier ce dernier dans la région du corps à traiter. Le principe est assez simple : le lance-pierre est chargé avec une molécule thérapeutique puis, une fois injecté, il circule dans l’organisme où il n’expulsera le médicament que lors de la rencontre avec un anticorps spécifique de la maladie à traiter.”
Radio-Canada– 2017-05-10
Un « lance-pierre » pour propulser des médicaments
“Une molécule en forme de lance-pierre faite d’ADN a été modélisée et synthétisée à l’échelle nanométrique par des chercheurs de l’Université de Montréal et leurs collègues de l’Université de Rome. Leur objectif est de propulser des médicaments à des endroits très précis du corps où se trouvent des biomarqueurs propres à une maladie. » Ce « lance-pierre moléculaire » est 20 000 fois plus petit qu’un cheveu humain et ne mesure que quelques nanomètres de long. Il se compose d’un brin d’ADN synthétique capable de transporter un médicament et de le relâcher au bon moment dans l’organisme à la manière de l’élastique d’un lance-pierre.”
The Engineer – 2017-05-08
Molecular “slingshot” releases drugs at specific targets
“A catapult-like microscopic machine made from DNA and enzymes could be used to deliver drug molecules to precise targets inside the body. The device, whose size is estimated at around a twenty-thousandth of the width of a hair, has been constructed by researchers at the University of Montréal and the University of Rome Tor Vergata. It comprises a length of synthetic DNA that can bind a drug molecule along its length, and terminates in anchoring groups that recognise and bind to the ends of Y-shaped antibody molecules in the blood stream. This DNA strand acts as the elastic in the catapult, while the antibody acts as the handle.”
Chemical & Engineering News – 2017-05-22
DNA slingshot’ targets drug delivery
“Gene therapy and personalized medicine are probably the first things that come to mind when people think about DNA and health care. But a growing number of researchers are viewing DNA as a functional and flexible material to build nanomachines with potential in medical diagnostics and drug delivery applications. The latest example is a “DNA slingshot” designed by researchers at the University of Montreal and the University of Rome Tor Vergata.”
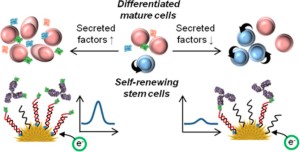
44. Zhou W, Mahshid SS, Wang W, Vallée-Bélisle A, Zandstra PW, Sargent EH, Kelley SO. “ Steric hindrance assay for secreted factors in stem cell culture” ACS Sens. 2(4):495−500. pdf
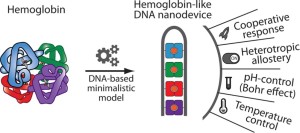
43. Mariottini D, Idili A, Vallée-Bélisle A, Plaxco KW, Ricci F. “A DNA nanodevice that loads and releases a cargo with hemoglobin-like allosteric control and cooperativity” Nano Lett. 17(5):3225–3230. pdf
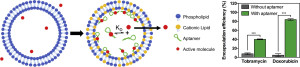
42. Plourde K, Derbali RM, Desrosiers A, Dubath C, Vallée-Bélisle A, Leblond J. “Aptamer-based liposomes improve specific drug loading and release” Journal of Controlled Release 251:82-91. pdf
2016
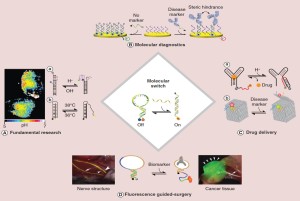
41. Desrosiers A, Vallée-Bélisle A*. “Nature-inspired DNA switches: applications in medicine” Nanomedicine (Lond). 12(3):175-179. pdf * corresponding author
40. Ricci F, Vallée-Bélisle A, Simon A, Porchetta A, Plaxco KW. “Using Nature’s “Tricks” To Rationally Tune the Binding Properties of Biomolecular Receptors” Acc. Chem. Res. 49(9):1884-92. pdf
39. Gareau D, Desrosiers A, Vallée-Bélisle A*. “Programmable Quantitative DNA Nanothermometers ” Nano Lett. 16(7):3976-81. pdf *corresponding author
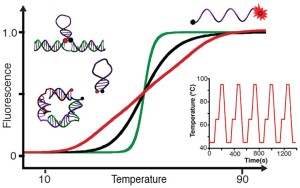
CBC-News– 2016-05-05
Montreal researcher builds world’s smallest thermometer
“Université de Montréal chemistry professor Alexis Vallée-Bélisle and his colleagues have spent the last three years building a nanothermometer so small, they can’t even see it. Vallée-Bélisle, who holds the Canada Research Chair in bioengineering and bio-nanotechnology, says he only knows it’s there because it’s working the way it was designed to. “The overall goal of the project was trying to build the smallest thermometer we could come up with. We know it’s there because it’s able to measure temperature exactly the way we want it to,” he told CBC’s Homerun. The thermometer is composed of structures made from DNA molecules. These structures are designed to auto-assemble since, at this scale, molecules are too small to be put together using nanotools.”
New York Post– 2016-04-27
Taking your temperature rectally will never be easier
“Scientists just created the world’s smallest thermometer — and it’s made from DNA. Chemists at the University of Montreal manipulated DNA molecules to create the programmable temperature checkers, which are 20,000 times smaller than a human hair. Their research, which was published Wednesday in the journal Nano Letters this week, will provide insight into how the body maintains the temperature of cells at a molecular level.”
Daily Mail (UK)– 2016-04-28
World’s tiniest thermometer made of DNA is 20,000 TIMES smaller than a human hair
“Biochemists have developed a thermometer from DNA that is 20,000 times smaller than a human hair. They designed the DNA strands to fold and unfold at different temperatures, allowing them to measure temperature changes between 86°F (30°C) and 185°F (85°C). They said such molecules could be used as nanoscale thermal switches or could also be used in combination to generate ultrasensitive thermometers.”
Yahoo Tech– 2016-04-29
Chemists create a DNA thermometer 20,000 times smaller than a human hair
“Did you know that DNA molecules unfold when heated? Scientists at the University of Montreal do, and they used that knowledge to design a thermometer made of DNA. It’s totally invisible to the naked eye, though, so it can be a bit difficult to read. Senior author of the paper “Programmable, quantitative, DNA Nanothermometers,” published in Nano Letters, Professor Alexis Vallée-Bélisle said, “Inspired by natural nanothermometers, which are typically 20,000x smaller than a human hair, we have created various DNA structures that can fold and unfold at specifically defined temperatures.” DNA is relatively easy to program, according to the scientists behind the project. There are four different nucleotides that bind differently to each other: the bond between A and T is weak, while C and G’s is strong. David Gareau, first author of the study, explained that “using simple design rules we are able to create DNA structures that fold and unfold at a specifically desired temperature.”
Metro– 2016-05-02
Nanotechnologie: le plus petit thermomètre au monde conçu à l’UdeM
“Mesurer la température à l’intérieur d’une seule cellule isolée d’un corps humain, est-ce possible? C’est ce que pourront tenter des chercheurs de l’Université de Montréal, qui ont construit un thermomètre à base d’ADN 20 000 fois plus petit qu’un cheveu humain, le plus petit modèle au monde. Cet instrument révolutionnaire permet de mesurer la température à une échelle nanométrique, une unité un million de fois plus petite qu’un millimètre. Il pourrait ouvrir la voie à de nombreuses applications dans le domaine médical, biologique et technologique, notamment dans la recherche contre le cancer.”
2015
38. Del Grosso E, Dallaire AM, Vallée-Bélisle A, Ricci F. “Enzyme-Operated DNA-Based Nanodevices ” Nano Lett. 15(12): 8407-11. pdf
37. Patskovsky S, Dallaire AM, Blanchard-Dionne AP, Vallée-Bélisle A, Meunier M. “Electrochemical structure-switching sensing using nanoplasmonic devices ” Ann. Phys. 527(11-12): 806–813. pdf
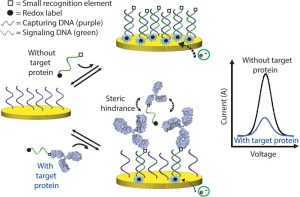
36. Mahshid SS, Camiré S, Ricci F, Vallée-Bélisle A*. “ A highly selective electrochemical DNA-based sensor that employs steric hindrance effects to detect proteins directly in whole blood ” J. Am. Chem. Soc. 137(50):15596-9. pdf *corresponding author
CBC-News– 2015-09-25
Montreal scientists score possible breakthrough for rapid diagnostic medical tests
“A simple and fast chemical process developed by scientists in Montreal could allow family doctors to one day use equipment as straightforward and cheap as a diabetic’s blood sugar tests to diagnose a range of diseases in minutes from their offices, instead of waiting days for results to come back from a lab.”
Genome Web– 2015-09-25
U of Montreal Team Develops Fast Electrochemical Sensor for Multiplexed Protein Marker Detection
“Researchers from the University of Montreal have developed a DNA-based electrochemical sensor that can detect the presence of multiple protein markers in whole blood in less than 10 minutes. The sensor could serve as the basis of a point-of-care device to diagnose a range of diseases and conditions including cancer, allergies, autoimmune diseases, and sexually transmitted diseases. “
TVA-Nouvelles– 2015-09-24
Les diagnostics médicaux à base d’ADN bientôt possibles à domicile
“Le diagnostic moléculaire, à base d’ADN, sera bientôt possible à domicile, selon des chimistes de l’Université de Montréal qui ont publié jeudi les résultats de leurs travaux dans le Journal of the American Chemical Society.”
Science Daily–2015-09-23
Molecular diagnostics at home: chemists design rapid, simple, inexpensive tests using DNA
“Chemists at the University of Montreal used DNA molecules to developed rapid, inexpensive medical diagnostic tests that take only a few minutes to perform. Their findings, which will officially be published tomorrow in the Journal of the American Chemical Society, may aid efforts to build point-of-care devices for quick medical diagnosis of various diseases ranging from cancer to allergies, autoimmune diseases, sexually transmitted diseases (STDs), and many others.”
35. Ranallo S, Rossetti M, Plaxco KW, Vallée-Bélisle A*, Ricci F * . “ A modular, DNA-based beacon for single-step fluorescence detection of antibodies and other proteins ” Angew. Chem., Int. Ed. Engl. 54: 1–6. pdf *co-corresponding authors
Chinese Edition of Scientific American – 2015-11-06
可以快速检测病毒的DNA纳米“机器”
“由合成DNA构成的纳米机器(nanoscalemachine ),可以快速、准确、廉价地诊断出包括艾滋病在内的多种疾病。”
Vice du jour – 2015-10-27 – Watch online video!
Détecter le VIH grâce à son téléphone
“Aujourd’hui… une innovation technologique permettrait de détecter le VIH avec un téléphone intelligent…”
La Presse Plus – 2015-10-19
Détecter le VIH grâce à son téléphone
“Diagnostiquer le VIH avec son téléphone intelligent à partir de chez soi ? Voilà le défi qu’une équipe de chercheurs de trois campus s’est lancé. Ils ont mis sur pied une nanomachine simplissime et efficace qui peut émettre sur-le-champ un diagnostic de VIH ou d’autres maladies auto-immunes et infectieuses. En s’inspirant de la mécanique naturelle de notre corps, les scientifiques ont renversé le modèle actuel, long et coûteux, des tests de dépistage. Incursion au cœur de l’infiniment petit qui voit grand.”
Wired – 2015-10-09
DNA-Nanomaschine kann HIV-Antikörper innerhalb von Minuten im Blut nachweisen
“Ein HIV-Test per Smartphone? Forscher haben jetzt eine DNA-Nanomaschine entwickelt, mit der sich eine HIV-Infektion binnen Minuten nachweisen lässt. Künftig soll der Test sogar auf Mobiltelefonen laufen können.”
Journal de Montréal – 2015-10-07
Des nanomachines pour détecter des maladies comme le VIH
“Une équipe de scientifiques a découvert une nouvelle approche qui pourrait permettre de développer des méthodes de détection rapides et abordables de maladies infectieuses et auto-immunes comme l’arthrite rhumatoïde et le VIH. L’équipe internationale, dont des chercheurs de l’Université de Montréal, a développé une nanomachine qui peut détecter et identifier des anticorps spécifiques.”
TVA-Nouvelles – 2015-10-07
Des nanomachines pour détecter des maladies graves
“Une équipe de scientifiques a découvert une nouvelle approche qui pourrait permettre de développer des méthodes de détection rapides et abordables de maladies infectieuses et auto-immunes comme l’arthrite rhumatoïde et le VIH. «Notre nanomachine offre d’importants avantages par rapport aux méthodes existantes de détection d’anticorps, a précisé le professeur Alexis Vallée-Bélisle de l’Université de Montréal, coauteur de l’étude. Rapide, elle ne nécessite pas de produits chimiques réactifs ni de manipulations complexes. Il s’agit simplement de l’ajouter à un échantillon sanguin et de mesurer l’intensité lumineuse produite”
Science Daily – 2015-10-07
Detecting HIV diagnostic antibodies with DNA nanomachines
“New research may revolutionize the slow, cumbersome and expensive process of detecting the antibodies that can help with the diagnosis of infectious and auto-immune diseases such as rheumatoid arthritis and HIV. An international team of researchers have designed and synthetized a nanometer-scale DNA “machine” whose customized modifications enable it to recognize a specific target antibody. Their new approach, which they described this month in Angewandte Chemie, promises to support the development of rapid, low-cost antibody detection at the point-of-care, eliminating the treatment initiation delays and increasing healthcare costs associated with current techniques.”
34. Idili A, Porchetta A, Vallée-Bélisle A*, Ricci F *. “ Controlling hybridization chain reactions with pH ” Nano Lett., 15 (8): 5539–44. pdf *co-corresponding authors
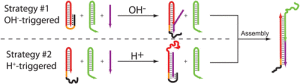
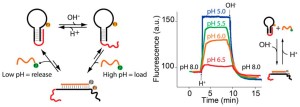
33. Porchetta A, Idili A, Vallée-Bélisle A, Ricci F. “ General strategy to introduce pH-induced allostery in DNA-based receptors to achieve controlled release of ligands” Nano Lett., 15(7):4467-71. pdf
32. Dallaire AM, Patskovsky S, Vallée-Bélisle A*, Meunier M *. “ Electrochemical plasmonic sensing system for highly selective multiplexed detection of biomolecules based on redox nanoswitches ” Biosens. Bioelectron. 71:75-81.pdf *co-corresponding authors
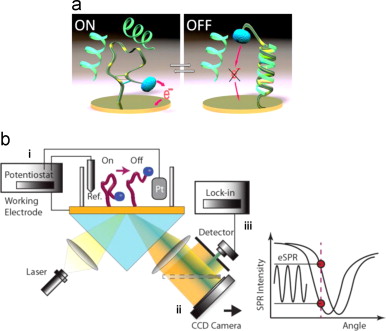
2014
31. Simon AJ, Vallée-Bélisle A, Ricci F, Watkins HM, Plaxco KW.. “ Using the population-shift mechanism to rationally introduce “Hill-type” cooperativity into a normally non-cooperative receptor. ” Angew. Chem., Int. Ed. Engl. 53(36):9471-5.pdf
30. Simon AJ, Vallée-Bélisle A, Ricci F, Plaxco KW.. “ Intrinsic disorder as a generalizable strategy for the rational design of highly responsive, allosterically cooperative receptors. ” Proc. Natl. Acad. Sci. U.S.A. 111(42):15048-53.pdf
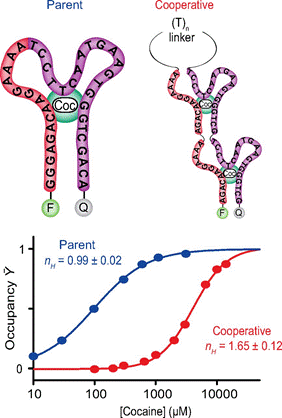
Science Magazine – 2014-11-21
Disorder enhances binding cooperativity
Some natural proteins, such as hemoglobin, bind their ligands in a cooperative manner, whereby the binding of the first ligand alters the structure of the protein, favoring binding of subsequent ligands. Taking inspiration from this mechanism, Simon et al. created unstructured receptors from DNA. Upon binding their ligand, the disordered receptors become structured, forming sites for further ligands to bind. Because ligands bind more favorably to these sites than to the unstructured receptor, the binding of subsequent ligands is enhanced. Relatively small changes in ligand concentration can therefore lead to an all-or-none binding response, a feature that could prove useful for artificial biotechnologies.
29. Idili A, Vallée-Bélisle A*, Ricci F*. “Programmable pH-triggered DNA nanoswitches. ” J. Am. Chem. Soc.136(16):5836-9. pdf *co-corresponding authors
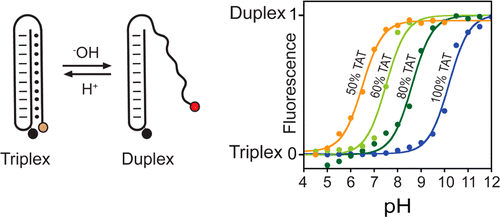
TVA Nouvelles – 2014-05-08
Nanodétecteurs : Le cancer traité par l’infiniment petit
Des bioingénieurs ont repoussé une fois de plus les limites de la science pour mettre au point des nanodétecteurs capables de détecter de minuscules variations chimiques, comme les changements de pH entre différentes cellules du corps humain. Cette technologie pourrait s’avérer utile dans le traitement de plusieurs maladies, dont le cancer, puisque les cellules cancéreuses ont généralement un pH inférieur à celui des cellules saines.
ScienceDaily – 2014-05-07
Using DNA to build tool that may literally shine light on cancer
Bioengineers at the University of Rome Tor Vergata and the University of Montreal have used DNA to develop a tool that detects and reacts to chemical changes caused by cancer cells and that may one day be used to deliver drugs to tumor cells.
2013
28. Idili A, Plaxco KW, Vallée-Bélisle A *, Ricci F *. “Thermodynamic basis for engineering high-affinity, high-specificity binding-induced DNA clamp nanoswitches.” ACS Nano. 7(12):10863-9. pdf *co-corresponding authors
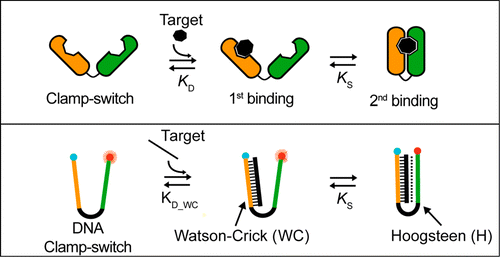
Science Daily – 2013-12-19
DNA Clamp to Grab Cancer Before It Develops
As part of an international research project, a team of researchers has developed a DNA clamp that can detect mutations at the DNA level with greater efficiency than methods currently in use. Their work could facilitate rapid screening of those diseases that have a genetic basis, such as cancer, and provide new tools for more advanced nanotechnology. The results of this research is published this month in the journal ACS Nano.
Medical Daily – 2013-12-19
New Nanotechnology Catches Cancer Mutations Before They Develop
Deleterious mutations could soon be spotted on a DNA level. In a new study from the University of California, Santa Barbara, nanotechnology developers describe their new “DNA clamp” — a groundbreaking innovation that will facilitate the detection of minute genetic variations associated with adverse health outcomes. The technology stands to revolutionize the treatment and diagnosis of certain cancers and other diseases with a genetic basis.
Montreal Gazette – 2013-12-27
‘DNA clamp’ technology may help screen for cancer
An international team of researchers has developed tiny technology that can spot genetic mutations at the DNA level before diseases such as cancer develop. Scientists are calling this nanotechnology a “DNA clamp” and say it could revolutionize screening and treatment of cancer, and other diseases with a genetic basis. “The clamp is made from DNA and it’s made to bind with DNA. It’s as small as it can get,” said Alexis Vallée-Bélisle, a professor in the chemistry department at the Université de Montréal, and one of the developers behind the discovery.
27. Lawrence C, Vallée-Bélisle A, Pfeil SH, de Mornay D, Lipman EA, Plaxco KW. “A comparison of the folding kinetics of a small, artificially selected DNA aptamer with those of equivalently simple naturally occurring proteins.” Protein Sci. 23(1):56-66. pdf
26. Porchetta A, Vallée-Bélisle A, Plaxco KW, Ricci F. “Allosterically tunable, DNA-based switches triggered by heavy metals.” J. Am. Chem. Soc. 135(36):13238-41. pdf
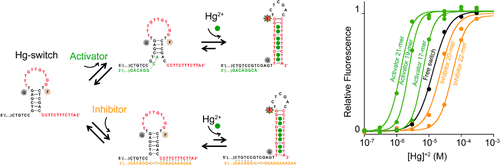
2012
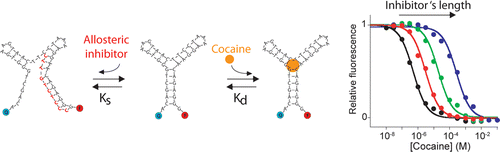
25. Porchetta A, Vallée-Bélisle A, Plaxco KW, Ricci F. “Using distal-site mutations and allosteric inhibition to tune, extend, and narrow the useful dynamic range of aptamer-based sensors.” J. Am. Chem. Soc. 134 20601-4. pdf
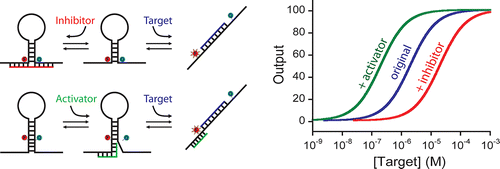
24. Ricci F, Vallée-Bélisle A, Porchetta A, Plaxco KW. “Rational Design of Allosteric Inhibitors and Activators Using the Population-Shift Model: In Vitro Validation and Application to an Artificial Biosensor” J. Am. Chem. Soc.134 15177-80. pdf
23. Vallée-Bélisle A, Ricci F, Uzawa T, Xia F, Plaxco KW. “Bioelectrochemical Switches for the Quantitative Detection of Antibodies Directly in Whole Blood” J. Am. Chem. Soc. 134 15197-200. pdf
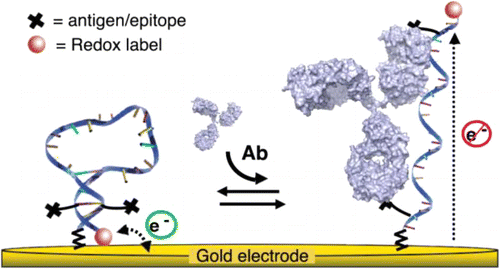
Science Daily – 2012-09-27
Bioengineers Design Rapid Diagnostic Tests Inspired by Nature
“By mimicking nature’s own sensing mechanisms, bioengineers at UC Santa Barbara and University of Rome Tor Vergata have designed inexpensive medical diagnostic tests that take only a few minutes to perform. Their findings may aid efforts to build point-of-care devices for quick medical diagnosis of sexually transmitted diseases (STDs), allergies, autoimmune diseases, and a number of other diseases….”
22. Vallée-Bélisle A & Michnick SW. “Visualizing transient protein-folding intermediates by tryptophan-scanning mutagenesis.” Nat. Struct. Mol. Biol. 19 731-736. pdf
Radio-Canada – 2012-06-13
Les Étapes de l’Assemblage des Protéines Visualisées
“Une nouvelle technique qui permet de visualiser l’assemblage des protéines, ces minuscules machines au coeur de la vie, a été mise au point par des bio-ingénieurs de l’Université de Montréal.” (voir p. 12)
Science Daily – 2012-06-10
Researchers Watch Tiny Living Machines Self-Assemble
“Enabling bioengineers to design new molecular machines for nanotechnology applications is one of the possible outcomes of a study by University of Montreal researchers that was published in Nature Structural and Molecular Biology.”
TechNewsWorld – 2012-06-13
Living Nanomachines Show Researchers the Ropes
“Nanomachines promise to transform the future of medicine. The fact is, though, our bodies already use them naturally. Two researchers have figured out a new way of observing and documenting what they do.”
Science Avenir – 2012-06-13
Observer la Formation des Protéines
“Une nouvelle méthode pour visualiser l’assemblage des protéines va permettre d’améliorer la compréhension de maladies causées par des erreurs d’ajustage comme Alzheimer ou Parkinson.”
Versions of this story were also printed in: Nanotechno.org, yahoo.com, World-science.net, Maxisciences.com, Techno-science.net, Agi.it, Corriere.it, Gen-es.org…
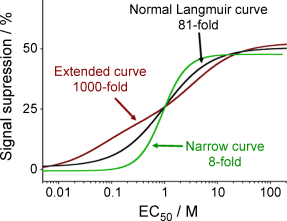
21. Kang D, Vallée-Bélisle A, Plaxco, KW, Ricci F. “Re-engineering Electrochemical Biosensors To Narrow or Extend Their Useful Dynamic Range” Angew. Chem., Int. Ed. Engl. 516717-21. pdf
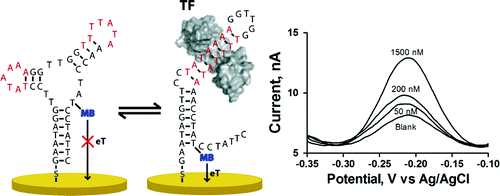
20. Bonham AJ, Hsieh K, Ferguson S, Vallée-Bélisle A, Ricci F, Plaxco KW. “Quantification of transcription factor binding in cell extracts using an electrochemical, structure-switching biosensor.” J. Am. Chem. Soc.,134 3346-8. pdf
19. Vallée-Bélisle A, Ricci F, Plaxco KW. “Engineering biosensors with extended, narrowed, or arbitrarily edited dynamic range” J. Am. Chem. Soc. 134 2876-9. pdf
Science Daily – 2012-02-14
Chemists mimic nature to design better medical tests
“Over their 3.8 billion years of evolution, living organisms have developed countless strategies for monitoring their surroundings. Chemists at UC Santa Barbara and University of Rome Tor Vergata have adapted some of these strategies to improve (…) DNA detectors.” (voir p.11)
Chemical & Engineering News – Vol. 90, Issue 2, January 9, 2012, p.9
Choose Your Own Analytical Range
“By combining sets of DNA receptors with different binding affinities, a research team has created biosensors that detect specific DNA sequences over customized concentration ranges.”
R & D Magazine – 2012-02-15
Chemists mimic nature to expand the range of biosensors
“Over their 3.8 billion years of evolution, living organisms have developed countless strategies for monitoring their surroundings. Chemists at UC Santa Barbara…”
Interviews and versions of this story were also printed in: The Daily Nexsus, The bottom line, Asknature.com, qmed.com, Nanotechnology.org, Sciencenewsline.com, Bio-medecine.org…
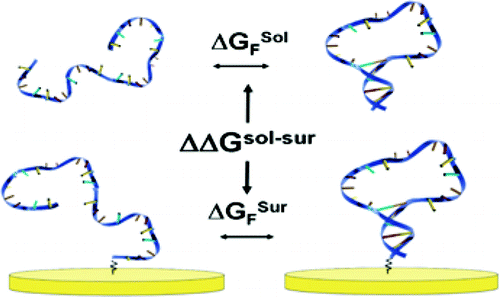
18. Watkins HM, Vallée-Bélisle A, Plaxco KW. “Entropic and Electrostatic Effects on the Folding Free Energy of a Surface-Attached Biomolecule: An Experimental and Theoretical Study” J. Am. Chem. Soc. 134 2120-6. pdf
17. Perez Rafael S, Vallée-Bélisle A, Fabregas E, Plaxco K, Palleschi G, Ricci F. “Employing the metabolic “branch point effect” to generate an all-or-none, digital-like response in enzymatic outputs and enzyme-based sensors.” Anal. Chem. 84 1076-82. pdf
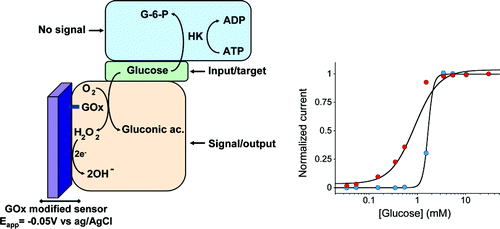
16. Kang D, White RJ, Vallée-Bélisle A, Plaxco KW, Xia F. “DNA-based biomolecular-electronic encoder and decoder devices constructed by multiplex biochip.” NPG Asian Materials 4 e1. pdf
2011
15. Ricci F, Vallée-Bélisle A, Plaxco KW. “High-Precision, in Vitro Validation of the Sequestration Mechanism for Generating Ultrasensitive Dose-Response Curves in Regulatory Networks” PLoS Comput Biol., 7 e1002171. pdf
14. Vallée-Bélisle A, Bonham AJ, Reigh NO, Ricci F, Plaxco KW. “Transcription factor beacons for the quantitative detection of DNA binding activities” J. Am. Chem. Soc. 133 13836-9. pdf
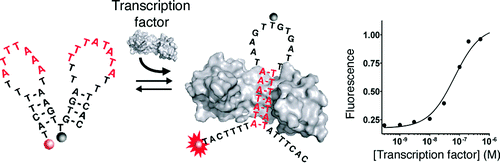
BioPhotonics, Photonics Media – Nov/Dec 2011, p.10
DNA nanosensors pave way for cancer tests, drugs
“DNA molecules can be used to make sensors that can quickly detect a broad class of proteins –and could personalize cancer treatment and even monitor the quality of stem cells…”
Santa Barbara News-Press – 2011-09-25, p.3
UCSB Bioengineers Light Path to New Cancer Tests
“You light up my life” may be more than just a song. Scientists at UCSB’s newly established Center for BioEngineering have designed molecules that could save lives by illuminating transcription factors…”
Science Daily – 2011-09-07
Nanosensors Made from DNA May Light Path to New Cancer Tests and Drugs
“Sensors made from custom DNA molecules could be used to personalize cancer treatments and monitor the quality of stem cells, according to an international team of researchers led by scientists at UC Santa Barbara and the University of Rome Tor Vergata.”
Versions of this story were also printed in: R & D Magazine, Sciencenewsline.com, Cancernewsdaily.com, Innovations-report.com, Physorg.com, Nanowerk.com, Materialstoday.com, Neurotransmitter.net, Biotechlive.com, Stemcellcafe.com…
2010
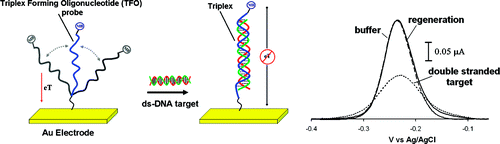
13. Patterson A, Caprio F, Vallée-Bélisle A, Moscone D, Plaxco KW, Palleschi G, Ricci F. “Using Triplex-Forming Oligonucleotide Probes for the Reagentless, Electrochemical Detection of Double-Stranded DNA.” Anal. Chem. 82 9109-15. pdf
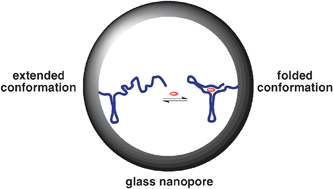
12. Abelow AE, Schepelina O, White RJ, Vallée-Bélisle A, Plaxco KW, Zharov I. “Biomimetic glass nanopores employing aptamer gates responsive to a small molecule.” Chem. Commun. 467984-6. pdf
11. Vallée-Bélisle A & Plaxco KW. “Structure-switching biosensors: inspired by Nature.” Curr. Opin. Struc. Biol. 20 518-26. pdf
10. Xia F, Zuo X, Yang R, Xiao Y, Kang D, Vallée-Bélisle A, Gong X, Yuen JD, Hsu BB, Heeger AJ , Plaxco KW. “Colorimetric detection of DNA, small molecules, proteins, and ions using unmodified gold nanoparticles and conjugated polyelectrolytes.” (2010) Proc. Natl. Acad. Sci. U.S.A. 9510837-41. pdf
9. Xia F, Zuo X, Yang R, White RJ, Xiao Y, Kang D, Gong X, Lubin AA, Vallée-Bélisle A, Yuen JD, Hsu BY, Plaxco KW. “Label-free, dual-analyte electrochemical biosensors: a new class of molecular-electronic logic gates.” J. Am. Chem. Soc. 132 8557-59. pdf
8. Xia F, Zuo X, Yang R, Xiao Y, Kang D, Vallée-Bélisle A, Gong X, Heeger AJ, Plaxco KW. “On the binding of cationic, water-soluble conjugated polymers to DNA: electrostatic and hydrophobic interactions.” J. Am. Chem. Soc. 132 1252-54. pdf
2009 and previous
7. Vallée-Bélisle A, Ricci F, Plaxco KW. (2009) “Thermodynamic basis for the optimization of binding-induced biomolecular switches and structure-switching biosensors.” Proc. Natl. Acad. Sci. U.S.A. 10613802-07. pdf
Science Daily – 2009-08-05
Chemists Explain The Switchboards In Our Cells
“Our cells are controlled by billions of molecular “switches” and chemists at UC Santa Barbara have developed a theory that explains how these molecules work…”.
Softpedia.com – 2009-08-04
Mathematical Model Finally Explains Cellular ‘Switches’
“Biologists at the University of California in Santa Barbara (UCSB) have recently developed a new model for explaining the actions of the billions of molecular ‘swiches’…”
R & D Magazine – 2009-08-10
Explaining the switchboards in our cells
“Our cells are controlled by billions of molecular “switches” and chemists at UC Santa Barbara have developed a theory that explains how these molecules work…”.
Versions of this story were also printed in Medical News Today, Chemistry times, World News, TopNews.com, ScientistLive.com, Littleabout.com, ScienceCentric.com, Chemie.de, Physorg.com, and RocketNews.com…
6. Vallée-Bélisle A. (2008) “Detection, characterization and visualization of protein transient states using tryptophane scanning.” Ph.D. Thesis, Université de Montréal, 1-232 (summa cum laude -highest honor)
5. Vallée-Bélisle A & Michnick SW (2007) “Multiple tryptophan probes reveal that ubiquitin folds via a late misfolded intermediate.” J. Mol. Biol. 374 791-805. pdf
4. Maxwell KL, Wildes D, Zarrine-Afsar A, De Los Rios MA, Brown AG, Friel CT, Hedberg L, Horng JC, Bona D, Miller EJ, Vallée-Bélisle A et al. (2005) “Protein folding: Defining a “standard” set of experimental conditions and a preliminary kinetic data set of two-state proteins.” Prot. Sci. 14602-616. pdf
3. Vallée-Bélisle A, Turcotte JF, Michnick SW (2004) “raf RBD and ubiquitin proteins share similar folds, folding rates and mechanisms despite having unrelated amino acid sequences.” Biochemistry 438447-8458. pdf
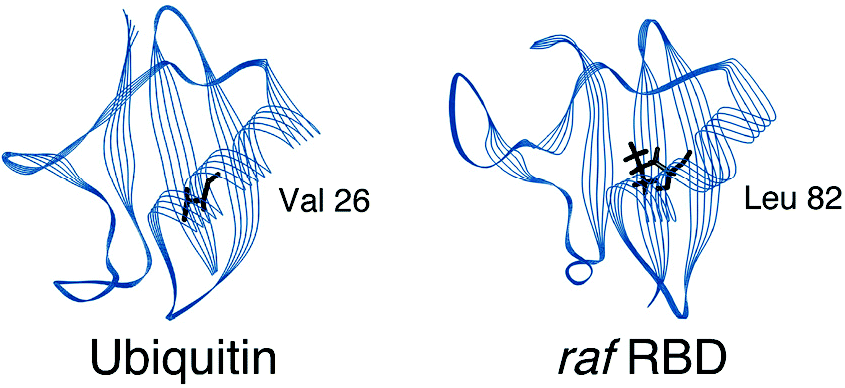
2. Vallée-Bélisle A (2000) “Le mécanisme de repliement de domain liant ras de raf: implication de la relation topologie-mécanisme de repliement pour les membres de la superfamille d’ubiquitine.” M. Sc. Thesis, Université de Montréal, 1-160
1. Michnick SW, Remy I, Campbell-Valois F–X, Vallée-Bélisle A, Pelletier JN (2000) “Detection of protein-protein interactions by protein fragment complementation strategies.” Methods Enzymol. 328 208-230
Prof. Alexis Vallée-Bélisle’s Patents
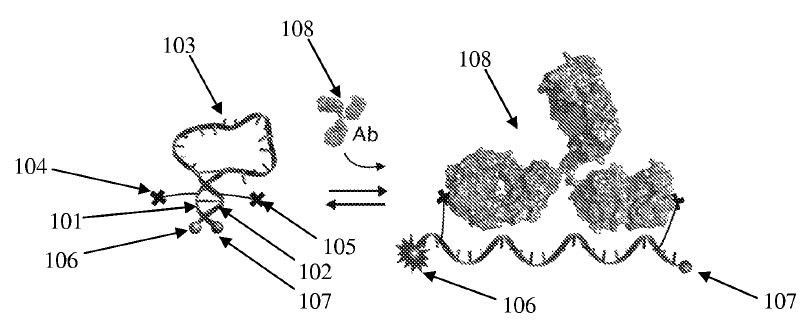
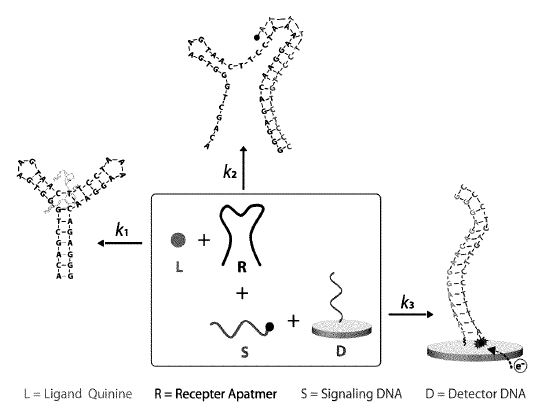
3. Vallée-Bélisle A, Guichi Z. “Kinetically programmed systems and reactions for molecular detection” US Patent 2021/0310057 A1, October 7, 2021. pdf
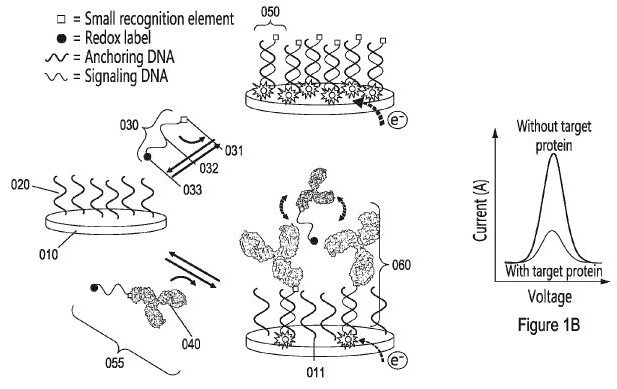
2. Vallée-Bélisle A, Mahshid SS. “Steric-hindrance hybridization systems, assays and methods associated thereto” US Patent 10,352,928 B2, July 16, 2019. pdf
1. Vallée-Bélisle A, Ricci F, White R, Bonham AJ, Plaxco KW. “Nucleotide-based probes and methods for the detection and quantification of macromolecules and other analytes” US Patent 9,828,628 B2, November 28, 2017. pdf